Home | About | Faculty | Calendar | Facilities | Graduate program | Contact | Apply
This page is optimized for viewing with javascript.
Brian W. Matthews

Emeritus, Physics
Member, IMB
Ph.D., University of Adelaide
D.Sc., University of Adelaide
B.S., University of Adelaide
Email
Office: Willamette Hall Room 376
Office Phone: 541-346-2572
Loading profile for Brian W. Matthews
Research Interests
Since his retirement Dr. Matthews does not have an active research group.
In the past our laboratory used X-ray crystallography, in concert with other techniques, to address some of the fundamental problems in biology: How do proteins spontaneously fold into their biologically active three-dimensional configurations? What determines the stability of these folded proteins? Can stability be improved? How do proteins interact with each other? How do proteins interact with DNA? How do enzymes interact with their substrates and act as catalysts?
We have used the lysozyme from bacteriophage T4 to define the contributions that different types of interaction make to the stability of proteins. One of the key findings is that the protein is, in general, very tolerant of amino acid replacement. This has permitted more challenging experiments such as the insertion or deletion of longer segments of the polypeptide chain. Such changes can be used to address a variety of questions regarding protein folding. It has recently become possible to monitor the behavior, including folding and catalysis, of single molecules. The wealth of information already available for T4 lysozyme makes it a very attractive subject for such studies and we are actively pursuing this new area.
Lysozymes with designed cavities are being used to test and to improve the effectiveness of docking programs designed to predict the optimal small-molecule that will bind to a given target site. Such sites are also being used to model the binding of general anesthetics.
We are also interested in the structural basis of DNA-protein interaction. Recent studies have focused on enzymes that are highly processive, i.e. they undergo multiple rounds of catalysis without dissociating from the substrate. In many, but not all cases, processivity can be achieved by having the enzyme completely enclose its substrate. In the case of lambda-exonuclease, for example, the enzyme forms a symmetrical toroid. For exonuclease I from E. coli, a toroid is also formed, but is by no means symmetrical (see figures).
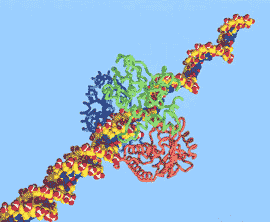
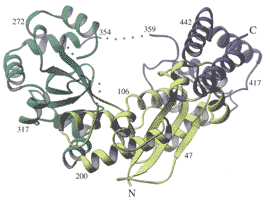 Model (top) showing the presumed mode by which
lambda-exonuclease encloses DNA and processively hydrolyzes
one of the two strands. The figure on the bottom shows the
structure of exonuclease I from E. coli. (Work of Rhett Kovall
and Wendy Breyer in the Matthews laboratory).
Model (top) showing the presumed mode by which
lambda-exonuclease encloses DNA and processively hydrolyzes
one of the two strands. The figure on the bottom shows the
structure of exonuclease I from E. coli. (Work of Rhett Kovall
and Wendy Breyer in the Matthews laboratory).
Several years ago we determined the three-dimensional structure of Escherichia coli beta-galactosidase, one of the classic enzymes in molecular biology. As well as studies of the enzyme, per se, we are also using this system to try to understand, in detail, the response of protein crystals to flash-freezing, an increasingly common step in contemporary X-ray crystallography.
Other areas of interest include structure-function studies of the F- and V-type ATPases, as well as various peptidases including the thermostable zinc protease thermolysin, the cobalt-requiring methionine aminopeptidase from E. coli as well as the serine peptidases.
Recent publications
(pulled from pubmed)
Recent publications
(pulled from pubmed)
Matthews BW
Protein Sci 2015 May;24(5):589-91
Matthews BW
Acta Crystallogr A 2013 Jan;69(Pt 1):34-6
Matthews BW
J Biomol Struct Dyn 2011 Feb;28(4):589-91; discussion 669-674
Baase WA, Liu L, Tronrud DE, Matthews BW
Protein Sci 2010 Apr;19(4):631-41
Liu L, Baase WA, Matthews BW
J Mol Biol 2009 Jan 16;385(2):595-605
Matthews BW, Liu L
Protein Sci 2009 Mar;18(3):494-502
Mooers BH, Tronrud DE, Matthews BW
Protein Sci 2009 May;18(5):863-70
Mooers BH, Baase WA, Wray JW, Matthews BW
Protein Sci 2009 May;18(5):871-80
Matthews BW
Protein Sci 2009 Jun;18(6):1135-8
Juers DH, Rob B, Dugdale ML, Rahimzadeh N, Giang C, Lee M, Matthews BW, Huber RE
Protein Sci 2009 Jun;18(6):1281-92
Liu L, Baase WA, Michael MM, Matthews BW
Biochemistry 2009 Sep 22;48(37):8842-51
Liu L, Marwitz AJ, Matthews BW, Liu SY
Angew Chem Int Ed Engl 2009;48(37):6817-9
Addlagatta A, Gay L, Matthews BW
Biochemistry 2008 May 13;47(19):5303-11
Liu L, Quillin ML, Matthews BW
Proc Natl Acad Sci U S A 2008 Sep 23;105(38):14406-11
Matthews BW
Protein Sci 2007 Jun;16(6):1013-6
Matthews BW
Nat Struct Mol Biol 2007 Jun;14(6):459-60
Juers DH, Lovelace J, Bellamy HD, Snell EH, Matthews BW, Borgstahl GE
Acta Crystallogr D Biol Crystallogr 2007 Nov;63(Pt 11):1139-53
Mooers BH, Matthews BW
Acta Crystallogr D Biol Crystallogr 2006 Feb;62(Pt 2):165-76
Wood ZA, Weaver LH, Brown PH, Beckett D, Matthews BW
J Mol Biol 2006 Mar 24;357(2):509-23
Sagermann M, Baase WA, Matthews BW
Protein Sci 2006 May;15(5):1085-92
Yousef MS, Bischoff N, Dyer CM, Baase WA, Matthews BW
Protein Sci 2006 Apr;15(4):853-61
Addlagatta A, Matthews BW
Protein Sci 2006 Aug;15(8):1842-8
Addlagatta A, Gay L, Matthews BW
Proc Natl Acad Sci U S A 2006 Sep 5;103(36):13339-44
Quillin ML, Wingfield PT, Matthews BW
Proc Natl Acad Sci U S A 2006 Dec 26;103(52):19749-53
Addlagatta A, Hu X, Liu JO, Matthews BW
Biochemistry 2005 Nov 15;44(45):14741-9
Juers DH, Kim J, Matthews BW, Sieburth SM
Biochemistry 2005 Dec 20;44(50):16524-8