Home | About | Faculty | Calendar | Facilities | Graduate program | Contact | Apply
This page is optimized for viewing with javascript.
S. James Remington

Professor, Physics
Member, IMB
Ph.D., University of Oregon
B.S., Oregon State University
Email
Office: Willamette Hall Room 377
Office Phone: 541-346-5190
Lab: Willamette Hall Room 354
Lab Phone: 541-346-5192
Loading profile for S. James Remington
Research Interests
Our group uses an interdisciplinary approach in applying physical techniques to the study of biological molecules, especially the structure, function, and interaction of enzymes, chemoreceptors and fluorescent proteins. The primary techniques we use are mutagenesis, x-ray crystallography and spectroscopy, but occasionally we perform computer modeling of enzyme active sites and other properties of proteins. In the laboratory, chemists and biologists collaborate with physicists to achieve a broader intellectual basis for the research.
Bacterial Chemoreceptor Proteins
In collaboration with Karen Guillemin’s group, we are interested to understand the structure and function of sensory proteins that allow the bacterium Helicobacter pylori to thrive within the hostile environment of the human stomach. Crystal structures were determined for TlpA and TlpB, two of the three critical chemotaxis receptors. Much subsequent work has, for the first time, led to a rather complete understanding of how a receptor (TlpB, see illustration) can sense pH, allowing the bacteria to navigate away from the low-pH interior of the stomach to the lining, where they can induce inflammation, ulcers and even cancerous transformations. Studies of TlpA, TlpB and TlpD are ongoing in both laboratories.
Green Fluorescent Protein
Since 1996 we have worked with Green Fluorescent Protein, which spontaneously rearranges itself to become fluorescent, absorbing blue light and re-emitting green light. GFP and its red, yellow and blue cousins are enormously popular as visible tag for proteins of interest or as a marker for gene expression. Using structure-based genetic engineering techniques we successfully constructed visual pH indicators, halide (chloride) concentration indicators and sensors that report on the thiol/disulfide redox potential within cells. Furthermore, the color of the protein can be modified by changing the environment or internal structure of the chromophore, which is derived from the primary sequence (Xaa)65-Tyr66-Gly67. Crystal structures were determined of related fluorescent proteins from corals that fluoresce yellow, orange and red, enabling multicolor reporting of a variety of cellular processes. It is fascinating that these different fluorescent proteins are nevertheless based on the same Xaa-Tyr-Gly peptide.
Enzyme Structure-Function Relationships
For many years we worked to determine structure function relationships in citrate synthase, which is the entry to the citric acid cycle and is found in every organism examined. Citrate synthase, in its rate-determining step, abstracts a proton from the methyl group of acetylCoenzyme A to form a carbon-carbon double bond. The side chain which accomplishes this task is Asp375 working in concert with His274 (sequence numbering of pig heart enzyme). This equilibrium for this seemingly simple reaction is disfavored in solution by 12-15 orders of magnitude, and proposals for how an enzyme can do this remain extremely controversial. Recently, we determined the crystal structure of malate synthase, an enzyme that catalyzes an essentially identical reaction. These enzymes are completely unrelated in sequence and structure, the underlying chemistry is essentially the same, but all of the details with the exception of an aspartic acid acting as a base are different. Evidently, Nature has discovered only one solution to this fundamental problem in chemistry, but the machinery is almost totally different!
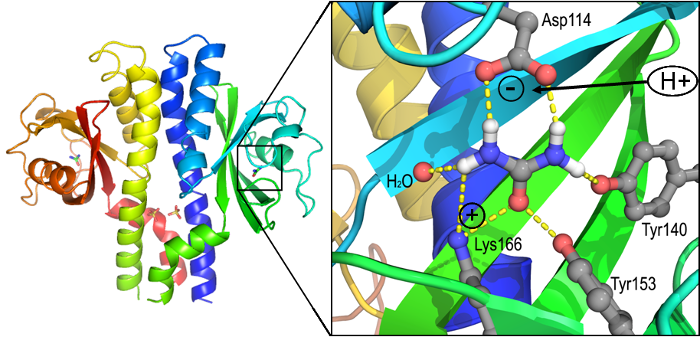 How TlpB senses acidic pH: The TlpB receptor (left) binds a molecule of
urea (right), which requires that Asp114 be negatively charged. At pH below
4, Asp114 becomes neutralized, which interferes with urea binding. This
in turn causes major structural changes in the receptor, which are
communicated through the transmembrane stalk to interior components.
Ultimately, the bacteria respond by swimming away from low pH (acidic)
environments, toward the stomach lining where they can cause great damage.
How TlpB senses acidic pH: The TlpB receptor (left) binds a molecule of
urea (right), which requires that Asp114 be negatively charged. At pH below
4, Asp114 becomes neutralized, which interferes with urea binding. This
in turn causes major structural changes in the receptor, which are
communicated through the transmembrane stalk to interior components.
Ultimately, the bacteria respond by swimming away from low pH (acidic)
environments, toward the stomach lining where they can cause great damage.
Recent publications
(pulled from pubmed)
Recent publications
(pulled from pubmed)
Anderson JK, Huang JY, Wreden C, Sweeney EG, Goers J, Remington SJ, Guillemin K
MBio 2015 Jul 7;6(4):e00379
Huang JY, Sweeney EG, Sigal M, Zhang HC, Remington SJ, Cantrell MA, Kuo CJ, Guillemin K, Amieva MR
Cell Host Microbe 2015 Aug 12;18(2):147-56
Schwarzländer M, Wagner S, Ermakova YG, Belousov VV, Radi R, Beckman JS, Buettner GR, Demaurex N, Duchen MR, Forman HJ, Fricker MD, Gems D, Halestrap AP, Halliwell B, Jakob U, Johnston IG, Jones NS, Logan DC, Morgan B, Müller FL, Nicholls DG, Remington SJ, Schumacker PT, Winterbourn CC, Sweetlove LJ, Meyer AJ, Dick TP, Murphy MP
Nature 2014 Oct 23;514(7523):E12-4
Goers Sweeney E, Henderson JN, Goers J, Wreden C, Hicks KG, Foster JK, Parthasarathy R, Remington SJ, Guillemin K
Structure 2012 Jul 3;20(7):1177-88
Shu X, Wang L, Colip L, Kallio K, Remington SJ
Protein Sci 2009 Feb;18(2):460-6
Henderson JN, Gepshtein R, Heenan JR, Kallio K, Huppert D, Remington SJ
J Am Chem Soc 2009 Apr 1;131(12):4176-7
Henderson JN, Osborn MF, Koon N, Gepshtein R, Huppert D, Remington SJ
J Am Chem Soc 2009 Sep 23;131(37):13212-3
Lohman JR, Remington SJ
Biochemistry 2008 Aug 19;47(33):8678-88
Lohman JR, Olson AC, Remington SJ
Protein Sci 2008 Nov;17(11):1935-45
Henderson JN, Ai HW, Campbell RE, Remington SJ
Proc Natl Acad Sci U S A 2007 Apr 17;104(16):6672-7
Shu X, Kallio K, Shi X, Abbyad P, Kanchanawong P, Childs W, Boxer SG, Remington SJ
Biochemistry 2007 Oct 30;46(43):12005-13
Shu X, Leiderman P, Gepshtein R, Smith NR, Kallio K, Huppert D, Remington SJ
Protein Sci 2007 Dec;16(12):2703-10
Cannon MB, Remington SJ
Protein Sci 2006 Jan;15(1):45-57
Henderson JN, Remington SJ
Physiology (Bethesda) 2006 Jun;21:162-70
Anstrom DM, Remington SJ
Protein Sci 2006 Aug;15(8):2002-7
Shu X, Shaner NC, Yarbrough CA, Tsien RY, Remington SJ
Biochemistry 2006 Aug 15;45(32):9639-47
Remington SJ
Curr Opin Struct Biol 2006 Dec;16(6):714-21
Remington SJ, Wachter RM, Yarbrough DK, Branchaud B, Anderson DC, Kallio K, Lukyanov KA
Biochemistry 2005 Jan 11;44(1):202-12
Quillin ML, Anstrom DM, Shu X, O'Leary S, Kallio K, Chudakov DM, Remington SJ
Biochemistry 2005 Apr 19;44(15):5774-87
Henderson JN, Remington SJ
Proc Natl Acad Sci U S A 2005 Sep 6;102(36):12712-7
Anstrom DM, Colip L, Moshofsky B, Hatcher E, Remington SJ
Acta Crystallogr Sect F Struct Biol Cryst Commun 2005 Dec 1;61(Pt 12):1069-74
Hanson GT, Aggeler R, Oglesbee D, Cannon M, Capaldi RA, Tsien RY, Remington SJ
J Biol Chem 2004 Mar 26;279(13):13044-53
Rossignol R, Gilkerson R, Aggeler R, Yamagata K, Remington SJ, Capaldi RA
Cancer Res 2004 Feb 1;64(3):985-93
Anstrom DM, Kallio K, Remington SJ
Protein Sci 2003 Sep;12(9):1822-32
Capaldi RA, Aggeler R, Gilkerson R, Hanson G, Knowles M, Marcus A, Margineantu D, Marusich M, Murray J, Oglesbee D, Remington SJ, Rossignol R
Biochim Biophys Acta 2002 Sep 10;1555(1-3):192-5
Hanson GT, McAnaney TB, Park ES, Rendell ME, Yarbrough DK, Chu S, Xi L, Boxer SG, Montrose MH, Remington SJ
Biochemistry 2002 Dec 31;41(52):15477-88
Yarbrough D, Wachter RM, Kallio K, Matz MV, Remington SJ
Proc Natl Acad Sci U S A 2001 Jan 16;98(2):462-7
Remington SJ
Methods Enzymol 2000;305:196-211
Wachter RM, Yarbrough D, Kallio K, Remington SJ
J Mol Biol 2000 Aug 4;301(1):157-71