Home | About | Faculty | Calendar | Facilities | Graduate program | Contact | Apply
This page is optimized for viewing with javascript.
Eric U. Selker

Professor, Biology
Member, IMB
Ph.D., Stanford University
B.S., Reed College
Email
Office: Streisinger Hall Room 355D
Office Phone: 541-346-5193
Lab: Streisinger Hall Room 355D
Lab Phone: 541-346-5197
Loading profile for Eric U. Selker
Research Interests
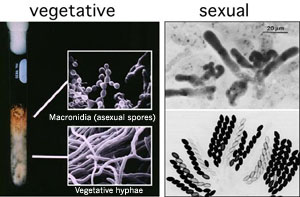
Figure 1. The filamentous fungus Neurospora crassa. Image courtesy of M. Springer and N. Raju
Overview
We are interested in how the eukaryotic genome is structured, how it functions, and how it changes. Our current research concentrates on gene silencing in eukaryotes. We are particularly interested in mechanisms involving special states of chromatin (e.g. heterochromatin) and DNA methylation. Methylation alters properties of DNA, affects DNA-protein interactions, represses genes in animals, plants, and fungi and is essential for normal development in plants and mammals. Remarkably little is understood, however, about what determines which chromosomal regions are methylated. We are using genetic and biochemical approaches, primarily with the filamentous fungus Neurospora crassa (Fig. 1) as a model, to elucidate the mechanism, regulation and function of DNA methylation. In addition, we are using these approaches to explore silencing associated with chromosome ends, centromeres and other specialized regions of the genome.
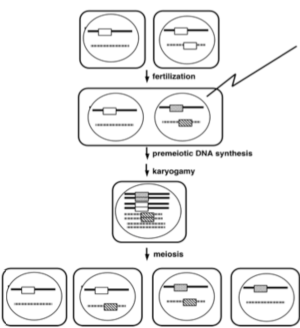
Figure 2. Inactivation of duplicated genes by RIP (shading) occurs prior to meiosis in Neurospora (see: Selker 1990 Ann. Rev. Gen. 24, 579-613).
DNA Methylation
DNA methylation is essential for normal development in a wide range of organisms including mammals and plants but is absent in some organisms including many popular model eukaryotes (e.g., yeasts, Drosophila, C. elegans). We showed that ~2% of cytosines in Neurospora DNA are methylated and that DNA methylation is not essential for development or viability in this organism. This set the stage for us to exploit this model eukaryote to elucidate the control and function of DNA methylation.
We found that most methylated regions of Neurospora are relics of transposons inactivated by RIP (repeat-induced point mutation), a premeiotic homology-based genome defense system that litters duplicated sequences with C:G to T:A mutations (Figs. 2 & 3).
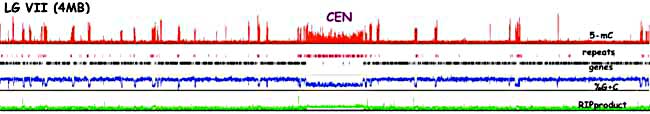
Figure 3. Detailed analyses of DNA methylation in the Neurospora genome revealed that it is mostly in AT-rich centromeric regions, subtelomeric regions and dispersed relics of RIP, as shown here in red for one chromosome (see Lewis et al. 2008 Genome Research 19, 427-437).
Our genetic and biochemical studies on the control of DNA methylation revealed clear ties between DNA methylation and chromatin modifications. The DIM-2 DNA methyltransferase is directed by heterochromatin protein 1 (HP1), which in turn recognizes trimethyl-lysine 9 on histone H3, placed by the DIM-5 histone H3 methyltransferase (Fig. 4).
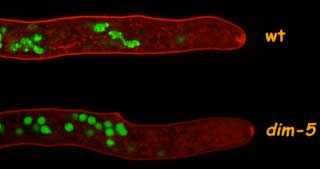
Figure 4. Localization of HP1-GFP depends on H3K9m3 by DIM-5. Note that the foci of heterochromatin in nuclei of wildtype are lost in the dim-5 mutant. (see Freitag M et. Al. 2004 Mol Cell 13, 427-34.)
DNA methylation is modulated by a variety of additional factors. For example, in dmm-1 (DNA methylation modulator-1) mutants, methylation spreads from inactivated transposable elements, which can silence adjacent genes and lead to poor growth. Additional studies in the laboratory are providing other insights into the workings of DNA methylation and other silencing processes.
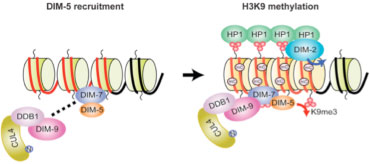
Figure 5. Heterochromatin formation and DNA methylation. DIM-7 recruits the DIM-5 histone methyltransferase to A:T rich DNA (red), to form DCDC (Dim-Cul4-DDB1 Complex) resulting in methylation of K9 of histone H3. HP-1 then recognizes this histone mark and recruits the DIM-2 DNA methyltransferase (see Lewis et al. 2010. PLoS Genetics 6, e1001196).
Group Members

Andy Klocko

Tereza Ormsby

Kevin McNaught

Jordan Gessaman

Vince Bicocca

Tish T. Wiles
Recent publications
(pulled from pubmed)
Recent publications
(pulled from pubmed)
Adhvaryu KK, Gessaman JD, Honda S, Lewis ZA, Grisafi PL, Selker EU
Eukaryot Cell 2015 Jan;14(1):25-8
Klocko AD, Rountree MR, Grisafi PL, Hays SM, Adhvaryu KK, Selker EU
PLoS Genet 2015 Mar;11(3):e1005083
Jamieson K, Rountree MR, Lewis ZA, Stajich JE, Selker EU
Proc Natl Acad Sci U S A 2013 Apr 9;110(15):6027-32
Honda S, Lewis ZA, Shimada K, Fischle W, Sack R, Selker EU
Nat Struct Mol Biol 2012 Apr 15;19(5):471-7, S1
Adhvaryu KK, Berge E, Tamaru H, Freitag M, Selker EU
PLoS Genet 2011 Dec;7(12):e1002423
Kothe GO, Kitamura M, Masutani M, Selker EU, Inoue H
Fungal Genet Biol 2010 Apr;47(4):297-309
Honda S, Lewis ZA, Huarte M, Cho LY, David LL, Shi Y, Selker EU
Genes Dev 2010 Mar 1;24(5):443-54
Lewis ZA, Adhvaryu KK, Honda S, Shiver AL, Selker EU
Proc Natl Acad Sci U S A 2010 May 4;107(18):8310-5
Rountree MR, Selker EU
Heredity (Edinb) 2010 Jul;105(1):38-44
Anderson DC, Green GR, Smith K, Selker EU
Biochemistry 2010 Jun 29;49(25):5244-57
Smith KM, Dobosy JR, Reifsnyder JE, Rountree MR, Anderson DC, Green GR, Selker EU
Genetics 2010 Dec;186(4):1207-16
Lewis ZA, Adhvaryu KK, Honda S, Shiver AL, Knip M, Sack R, Selker EU
PLoS Genet 2010 Nov 4;6(11):e1001196
Lewis ZA, Honda S, Khlafallah TK, Jeffress JK, Freitag M, Mohn F, Schübeler D, Selker EU
Genome Res 2009 Mar;19(3):427-37
Wu C, Kim YS, Smith KM, Li W, Hood HM, Staben C, Selker EU, Sachs MS, Farman ML
Genetics 2009 Mar;181(3):1129-45
Honda S, Selker EU
Genetics 2009 May;182(1):11-23
Selker EU
Genetics 2008 Feb;178(2):611-9
Honda S, Selker EU
Mol Cell Biol 2008 Oct;28(19):6044-55
Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA, Johnson EA
PLoS One 2008;3(10):e3376
Smith KM, Kothe GO, Matsen CB, Khlafallah TK, Adhvaryu KK, Hemphill M, Freitag M, Motamedi MR, Selker EU
Epigenetics Chromatin 2008 Nov 3;1(1):5
Adhvaryu KK, Selker EU
Genes Dev 2008 Dec 15;22(24):3391-6
Lewis ZA, Shiver AL, Stiffler N, Miller MR, Johnson EA, Selker EU
Genetics 2007 Oct;177(2):1163-71
Freitag M, Selker EU
Curr Opin Genet Dev 2005 Apr;15(2):191-9
Adhvaryu KK, Morris SA, Strahl BD, Selker EU
Eukaryot Cell 2005 Aug;4(8):1455-64
Freitag M, Hickey PC, Khlafallah TK, Read ND, Selker EU
Mol Cell 2004 Feb 13;13(3):427-34
Freitag M, Lee DW, Kothe GO, Pratt RJ, Aramayo R, Selker EU
Science 2004 Jun 25;304(5679):1939
Freitag M, Hickey PC, Raju NB, Selker EU, Read ND
Fungal Genet Biol 2004 Oct;41(10):897-910
Selker EU
Cold Spring Harb Symp Quant Biol 2004;69:119-24
Tamaru H, Zhang X, McMillen D, Singh PB, Nakayama J, Grewal SI, Allis CD, Cheng X, Selker EU
Nat Genet 2003 May;34(1):75-9
Selker EU, Tountas NA, Cross SH, Margolin BS, Murphy JG, Bird AP, Freitag M
Nature 2003 Apr 24;422(6934):893-7