Home | About | Faculty | Calendar | Facilities | Graduate program | Contact | Apply
This page is optimized for viewing with javascript.
Ken Prehoda

Professor, Chemistry and Biochemistry
Member, IMB
Ph.D., University of Wisconsin
B.A., California State University
Email
Lab website
Office: Klamath Hall Room 291
Office Phone: 541-346-5030
Lab: Klamath Hall Room 290
Lab Phone: 541-346-5049
Loading profile for Ken Prehoda
Research Interests
Research in the Prehoda lab focuses on understanding the protein interaction networks that regulate stem cell divisions. The Drosophila neuroblast undergoes repeated asymmetric divisions to populate the fly central nervous system and is our primary model system. Following neuroblast division, one daughter cell retains the neuroblast fate while the other goes on to become neurons or glia. The foundation of this amazing process is the segregation of different fate determinant proteins into the two daughter cells during mitosis. Our work attempts to uncover the molecular basis of this stem cell division through the study of cell polarity and mitotic spindle orientation.
The first step in fate determinant segregation is their polarization, in which the proteins localize to opposite regions of the cell membrane. How does the cell become organized in this way? We have studied the Par complex, a set of three proteins that polarizes many different cell types, including neuroblasts. A kinase, atypical Protein Kinase C, is a key component of the Par complex, as attachment of a phosphate to downstream proteins polarizes them. We are trying to understand how cells control where the Par complex is localized, and how substrate phosphorylation leads to their polarization. Loss of polarity is a hallmark of cancer, and we are also studying the relationship between polarity and tumorigenesis.
Fate determinant polarization alone is not enough for the stem cell to function properly. The mitotic spindle must also align along the polarity axis so that the cleavage furrow will bisect the the two membrane domains containing different fate determinants. How is the position of the spindle specified? We are studying proteins that link the cell cortex with microtubule motors that generate forces on the spindle's astral microtubules.
We have also recently begun working on questions in molecular evolution. One of the spindle orienting proteins we've identified is a protein interaction domain that resembles the guanylate kinase (GK) enzyme, which catalyzes the formation of GDP from GMP and ATP. The GK domain version of the protein originated via a duplication of the enzyme sometime near the unicellular to multicellular transition that gave rise to animals (approximately 600 million years ago). How did a nucleotide kinase become a protein interaction domain? In collaboration with the Thornton and Harms labs, we have "resurrected" the ancient ancestors of these proteins so that they can be studied in the lab. This has allowed us to directly answer questions about this amazing functional transition. As the GK domain is a special type of protein interaction domain - a phosphoprotein recognition module - we are now studying how regulation evolved in this system.
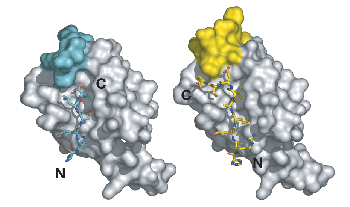 Structures of PDZ domain complexes involved in stem cell division. On the left, the PDZ from the protein Par-6 is bound to a carboxy-terminal sequence (N and C-terminii of the ligand are labeled). This mode of binding is enforced by a steric mechanism residues that would extend past the c-terminus would clash with residues from the domain itself. On the right, an internal sequence is shown bound to the Par-6 PDZ domain. This sequence bypasses the carboxy-terminal requirement by taking advantage of plasticity in the PDZ domain.
Structures of PDZ domain complexes involved in stem cell division. On the left, the PDZ from the protein Par-6 is bound to a carboxy-terminal sequence (N and C-terminii of the ligand are labeled). This mode of binding is enforced by a steric mechanism residues that would extend past the c-terminus would clash with residues from the domain itself. On the right, an internal sequence is shown bound to the Par-6 PDZ domain. This sequence bypasses the carboxy-terminal requirement by taking advantage of plasticity in the PDZ domain.
Recent publications
(pulled from pubmed)
Recent publications
(pulled from pubmed)
Graybill C, Prehoda KE
Biochemistry 2014 Aug 5;53(30):4931-7
Johnston CA, Manning L, Lu MS, Golub O, Doe CQ, Prehoda KE
J Cell Sci 2013 Oct 1;126(Pt 19):4436-44
Lu MS, Prehoda KE
Dev Cell 2013 Aug 26;26(4):369-80
Mauser JF, Prehoda KE
PLoS One 2012;7(1):e29611
Graybill C, Wee B, Atwood SX, Prehoda KE
J Biol Chem 2012 Jun 15;287(25):21003-11
Johnston CA, Doe CQ, Prehoda KE
PLoS One 2012;7(4):e36014
Lu MS, Mauser JF, Prehoda KE
ACS Synth Biol 2012 Feb 17;1(2):65-72
Smith NR, Prehoda KE
Mol Cell 2011 Aug 19;43(4):540-9
Connell M, Cabernard C, Ricketson D, Doe CQ, Prehoda KE
Mol Biol Cell 2011 Nov;22(22):4220-6
Johnston CA, Whitney DS, Volkman BF, Doe CQ, Prehoda KE
Proc Natl Acad Sci U S A 2011 Nov 1;108(44):E973-8
Wee B, Johnston CA, Prehoda KE, Doe CQ
J Cell Biol 2011 Oct 31;195(3):369-76
Cabernard C, Prehoda KE, Doe CQ
Nature 2010 Sep 2;467(7311):91-4
Ricketson D, Johnston CA, Prehoda KE
Proc Natl Acad Sci U S A 2010 Dec 7;107(49):20964-9
Newman RA, Prehoda KE
J Biol Chem 2009 May 8;284(19):12924-32
Atwood SX, Prehoda KE
Curr Biol 2009 May 12;19(9):723-9
Johnston CA, Hirono K, Prehoda KE, Doe CQ
Cell 2009 Sep 18;138(6):1150-63
Marcette J, Hood IV, Johnston CA, Doe CQ, Prehoda KE
Biochemistry 2009 Oct 27;48(42):10014-9
Prehoda KE
Cold Spring Harb Perspect Biol 2009 Aug;1(2):a001388
Liu SL, Fewkes N, Ricketson D, Penkert RR, Prehoda KE
J Biol Chem 2008 Jan 4;283(1):380-7
Atwood SX, Chabu C, Penkert RR, Doe CQ, Prehoda KE
J Cell Sci 2007 Sep 15;120(Pt 18):3200-6
Nipper RW, Siller KH, Smith NR, Doe CQ, Prehoda KE
Proc Natl Acad Sci U S A 2007 Sep 4;104(36):14306-11
Qian Y, Prehoda KE
J Biol Chem 2006 Nov 24;281(47):35757-63
Penkert RR, DiVittorio HM, Prehoda KE
Nat Struct Mol Biol 2004 Nov;11(11):1122-7
Prehoda KE, Lim WA
Curr Opin Cell Biol 2002 Apr;14(2):149-54